How Arp2/3 Complex Nucleates Actin Filaments
Background: Arp2/3 and actin-based motility
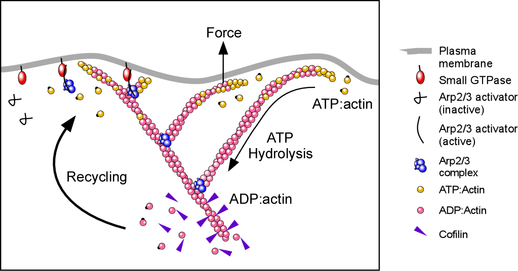
The Arp2/3 complex builds crosslinked actin networks that drive cell movement, making it possible for immune cells to catch bacteria, or neurons migrate and wire up to the right place in the brain. Arp2/3 builds these networks by binding to the sides of existing actin filaments and nucleates growth of new filaments from their sides, resulting in a dendritic filament array.
To understand how Arp2/3 nucleates filaments, we focused in on the Arp2 and Arp3 subunits of the 7-subunit protein complex. These Arps (Actin Related Proteins) have a strikingly similar structure to actin, including the ATP binding pocket, so we looked to see if they bound and hydrolysed ATP.
ATP binding by Arp2/3
We measured the binding kinetics of Arp2/3 to ATP, and found that both Arp2 and Arp3 bind ATP, but which much weaker affinity than actin binds ATP. This allowed us to use a kinetic trick—loading actin with ATP, but keeping the ATP concentration very low then mixing with a large amount of ADP, (or AMP-PNP, a non-hydrolyzable analogue of ATP) just before starting a nucleation experiment. In this way, we could see if changing the nucleotide on Arp2/3 changed its ability to nucleate, while keeping ATP on actin. We found that Arp2/3 needs ATP (and not ADP or AMP-PNP) to nucleate new filaments, suggesting that it might be hydrolyzed during nucleation.
Looking for ATP hydrolysis on Arp2/3 is difficult, because when activated Arp2/3 nucleates actin polymerization, and actin itself hydrolyses ATP when it polymerizes. The challenge is to measure ATP hydrolysis on minute (nM) concentrations of Arp2/3 complex in a solution containing large (uM) concentrations of ATP-hydrolyzing actin.
ATP hydrolysis by Arp2/3
The trick we used to measure ATP hydrolysis on Arp2/3 was to replace normal ATP with a radio-labeled version (32P on the Ɣ-phosphate), and crosslink this covalently to the Arp2 and Arp3 subunits of the complex. We washed it so that the only 32P in the reaction was on the Arp2/3, so we could then run the samples out on a gel to separate the proteins, and look for loss of the 32P labeling over time (which indicates cleavage of the Ɣ-phosphate and ATP hydrolysis). Because the only 32P-ATP in the reaction is physically attached to the Arp2/3 complex, we don’t have to worry about ATP hydrolysis by actin confounding the experiment. When we performed a nucleation reaction with this labeled Arp2/3, we found that Arp2 rapidly hydrolyses ATP when Arp2/3 nucleates new filaments, and we didn’t see any hydrolysis of ATP on Arp3.
Investigating the Mechanism of Filament Nucleation
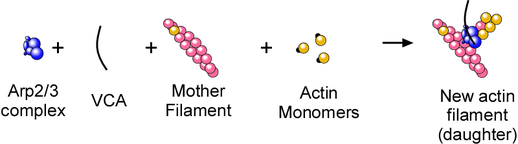
The fact that ATP hydrolysis occurs rapidly on Arp2 upon nucleation meant we could use ATP hydrolysis as a probe for activation of the Arp2/3 complex. These are the components of the nucleation reaction:
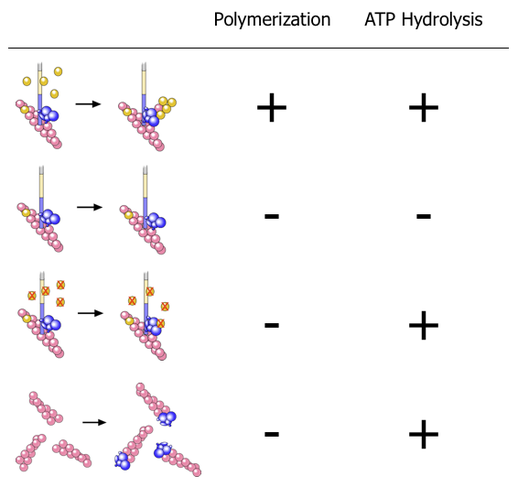
We put all of the factors required to nucleate actin filaments (monomeric actin, actin filaments, and a nucleating factor(VCA)) in various combinations to see if they activated ATP hydrolysis on Arp2 (using Latrunculin A and Phalloidin to keep the monomers monomeric and the polymers from depolymerizing.) Here’s what we found:
- The first line is the normal polymerization reaction—nothing surprising there.
- The second line shows that without actin monomers, there’s no ATP hydrolysis on Arp2.
- The third line shows that adding actin monomers does trigger ATP hydrolysis on Arp2 even when those monomers are prevented from polymerizing by Latrunculin A—i.e. one monomer is all that’s required to trigger Arp2 to hydrolyze ATP (in the presence of the other components), you don’t need polymerization.
- The fourth line is particularly interesting. Here we took Arp2/3, mixed it with filaments then sheared the filaments to break them. Arp2/3 then attaches to the ends of the filaments (capping), much like it is attached to the end of the new filament it nucleates when it forms branches. This capping also triggers Arp2 to hydrolyse ATP, but without the other components (the mother filament and VCA)!
This is explained in more detail in the paper, but the bottom line is that it explains how Arp2/3 is working, implying that Arp2 is behaving like an actin monomer does in a filament.
How we think Arp2/3 nucleates new actin filaments
When actin monomers polymerize to make a filament, the protein conformation of the monomer changes and causes it to hydrolyze ATP. Here’s a model showing this, with the conformational change shown by the little red levers that extend out to contact the core of the filament (actin shown in white):
Model for how a conformational change in actin within a filament triggers ATP hydrolysis
The idea is that Arp2 is doing the same thing, but that the Arp2 is put in a filament-like conformation by the assembly of a nucleus (Arp2-Arp3-monomer). The clearest evidence for this is the trigger of ATP hydrolysis by capping, which directly creates the Arp2-Arp3-monomer arrangement (actin shown in white, Arp2 in blue, Arp3 in pink):

Model for how Arp2/3 docks onto the pointed end of an actin filament, putting Arp2 and Arp3 into a filament-like conformation and triggering ATP hydrolysis on Arp2
And the Latrunculin result (that when filaments and VCA are present, you just need one monomer) implies that when nucleation is triggered by the formation of this Arp2-Arp3-monomer nucleus, with VCA bringing in the monomer.
Putting this together gives us the model for filament nucleation (actin shown in white, Arp2 in blue, Arp3 in pink):

Model for how Arp2/3 nucleates actin filaments: VCA brings an actin monomer to the Arp2/3 complex, creating an Arp2-Arp3-actin nucleus and triggering ATP hydrolysis on Arp2. Actin polymerizes from this nucleus to form the daughter filament.
Function of ATP hydrolysis
We used ATP hydrolysis to probe the nucleation mechanism, and didn’t directly address the function of ATP hydrolysis itself. It may be, like actin, involved with later disassembly events (for actin this is really determined by phosphate (Pi) release, which is downstream of, and occurs much later thanm ATP hydrolysis).
References
[1] Activation of Arp2/3 complex: addition of the first subunit of the new filament by a WASP protein triggers rapid ATP hydrolysis on Arp2.
Dayel MJ, Mullins RD.
PLoS Biol. 2004 Apr;2(4):E91. Epub 2004 Apr 13.
PMID: 15094799
Free Open-access article
[2] Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments.
Dayel MJ, Holleran EA, Mullins RD.
Proc Natl Acad Sci U S A. 2001 Dec 18;98(26):14871-6.
PMID: 11752435
Free Open-access article