CometWiki
CometWiki
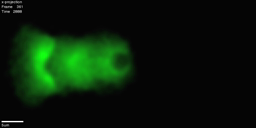
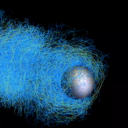
IntroductionThis site describes the set up and usage of `comet', an actin-based bead motility simulator. Cells in our body use Actin to move. Unlike the actin-myosin interaction that produces muscle movement, this is on the scale of individual cells and allows, for example, neurons to migrate and wire up to the right part of the brain, and cells in our immune system to track down and engulf bacteria. To achieve this movement, cells lay down actin polymer networks that produce force. One way to study how our cells use actin to produce force and move is to study a simplified system which still recreates actin-based motility in vitro, such as 'actin-based bead motility'. In actin-based bead motility, we coat a bead with proteins that tell the cell to polymerize actin, then put the bead a solution similar to that inside of the cell, which causes an actin network to build around the bead. Surprisingly, even when the bead is spherically symmetric, rather than just building a symmetric shell that gets bigger and bigger, it moves off in 'comet tail' of actin. This bead motility simulator aims to help us understand how this process works. Simulator OutputExample Results
Model RobustnessIn Vitro
Essential InformationInstalling the program
Running the programThe program is called from the command line. The command line parameters tell the program what to do (calculate a new run, re-process existing data, interactive 3D view etc.). A
How the program works
In Depth InformationMaking Measurments
|
Code Status |